Abstract
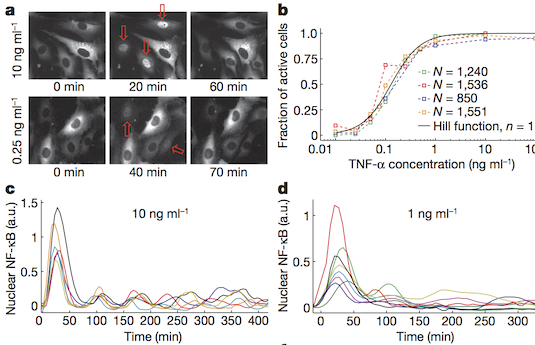
Cells operate in dynamic environments using extraordinary communication capabilities that emerge from the interactions of genetic circuitry. The mammalian immune response is a striking example of the coordination of different cell types. Cell-to-cell communication is primarily mediated by signalling molecules that form spatiotemporal concentration gradients, requiring cells to respond to a wide range of signal intensities. Here we use high-throughput microfluidic cell culture and fluorescence microscopy, quantitative gene expression analysis and mathematical modelling to investigate how single mammalian cells respond to different concentrations of the signalling molecule tumour-necrosis factor (TNF)-α, and relay information to the gene expression programs by means of the transcription factor nuclear factor (NF)-κB. We measured NF-κB activity in thousands of live cells under TNF-α doses covering four orders of magnitude. We find, in contrast to population-level studies with bulk assays, that the activation is heterogeneous and is a digital process at the single-cell level with fewer cells responding at lower doses. Cells also encode a subtle set of analogue parameters to modulate the outcome; these parameters include NF-κB peak intensity, response time and number of oscillations. We developed a stochastic mathematical model that reproduces both the digital and analogue dynamics as well as most gene expression profiles at all measured conditions, constituting a broadly applicable model for TNF-α-induced NF-κB signalling in various types of cells. These results highlight the value of high-throughput quantitative measurements with single-cell resolution in understanding how biological systems operate.
Where applicable, full text and supplement provided for fair use.