Abstract
Introduction: Logistic regression models are used in genomic studies to analyze the genetic data linked to electronic health records (EHRs), and do not take full usage of the time-to-event information available in EHRs. Previous work has shown that Cox regression, which can account for left truncation and right censoring in EHRs, increased the power to detect genotype-phenotype associations compared to logistic regression. We extend this to evaluate the relative performance of Cox regression and various logistic regression models in the presence of positive errors in event time (delayed event time), relating to recorded event time accuracy.
Methods: One Cox model and three logistic regression models were considered under different scenarios of delayed event time. Extensive simulations and a genomic study application were used to evaluate the impact of delayed event time.
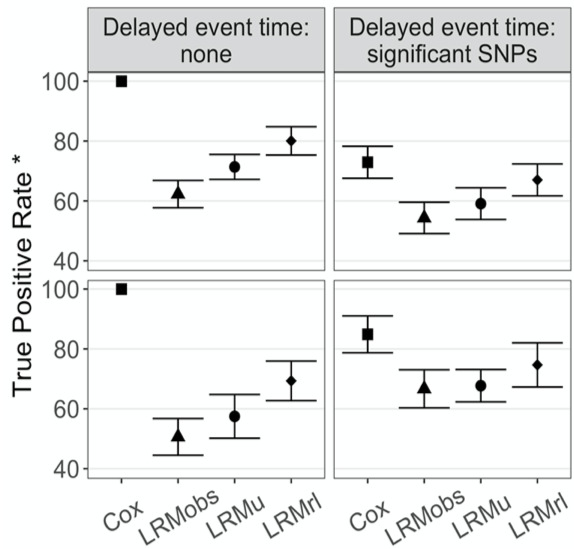
Results: While logistic regression does not model the time-to-event directly, various logistic regression models used in the literature were more sensitive to delayed event time than Cox regression. Results highlighted the importance to identify and exclude the patients diagnosed before entry time. Cox regression had similar or modest improvement in statistical power over various logistic regression models at controlled type I error. This was supported by the empirical data, where the Cox models steadily had the highest sensitivity to detect known genotype-phenotype associations under all scenarios of delayed event time.
Conclusion: With or without the presence of delayed event time scenarios that might exist in EHRs, Cox regression outperformed the logistic regression models commonly used in genomic studies.
Where applicable, full text and supplement provided for fair use.